Preliminary Mapping of Soybean Dominant Locus Hrcs7 Conferring Resistance to Cerocospara sojina Race 7 
 Author
Author  Correspondence author
Correspondence author
Molecular Plant Breeding, 2011, Vol. 2, No. 6 doi: 10.5376/mpb.2011.02.0006
Received: 29 Dec., 2010 Accepted: 25 Feb., 2011 Published: 15 Apr., 2011
Dong et al., 2011, Preliminary mapping of soybean dominant locus Hrcs7 conferring resistance to Cerocospara sojina race 7, Molecular Plant Breeding Vol.2 No.6 (doi: 10.5376/mpb.2011.02.0006)
Molecular marker-assisted selection can significantly improved resistance breeding efficiency. The soybean frogeye leaf spot (FLS) is caused by the fungus Cercospora sojina Hara. In order to find the resistant markers linked to C. sojina race 7 in soybean, we carried out the resistant genetic ananlysis and the resistant locus mapping. The F2 population derived from the cross of 'Gang 95144-1' and 'Gongjiao 9723-6' was employed to analyze the resistance genetics. it was sure that the resistance conferring to FLS was controlled by one major dominant gene.. Bulk Segregant Analysis (BSA) and Simple Sequence Repeat (SSR) method were used to map the resistant locus. Among the 600 selected SSR primers, there were four primer pairs exhibiting the polymorphisms between DNA bulk of resistant and susceptible as well as both parents. Segregation distribution of the SSR markers, satt 414 and satt384, were conformed significantly to be as 1:2:1.of Mendel Segregation. The marker, satt411 and satt384 were assigned to link with the resistant locus based on estimation of MAPMAKER/EXP 3.0 software. The linkage order and distance would be Satt384 -5.7cM - Satt411 -7.9cM -Hrcs7. The resistant locus was integrated to soybean linkage group E based on the Cregen's genetic linkage map of soybean.Molecular marker-assisted selection can significantly improved resistance breeding efficiency. The soybean frogeye leaf spot (FLS) is caused by the fungus Cercospora sojina Hara. In order to find the resistant markers linked to C. sojina race 7 in soybean, we carried out the resistant genetic ananlysis and the resistant locus mapping. The F2 population derived from the cross of 'Gang 95144-1' and 'Gongjiao 9723-6' was employed to analyze the resistance genetics. it was sure that the resistance conferring to FLS was controlled by one major dominant locus named Hrcs7.. Bulk Segregant Analysis (BSA) and Simple Sequence Repeat (SSR) method were used to map the resistant locus. Among the 600 selected SSR primers, there were four primer pairs exhibiting the polymorphisms between DNA bulk of resistant and susceptible as well as both parents. Segregation distribution of the SSR markers, satt 414 and satt384, were conformed significantly to be as 1:2:1.of Mendel Segregation. The marker, satt411 and satt384, were assigned to link with the resistant locus based on estimation of MAPMAKER/EXP 3.0 software. The linkage order and distance would be Satt384 -5.7cM -Satt411 -7.9cM -Hrcs7. The resistant locus was integrated to soybean linkage group E based on the Cregen's genetic linkage map of soybean.
Frogeye leaf spot (FLS) is an important disease of soybean in the world for several years (Yorinori, 1997). It is caused by Cercospora sojina K. Hara. There are several races of this fungal pathogen, which are distinguished by their ability to infect soybean varieties. There were 12 races in America, 20 races in Brazil, 11 races in China. But only China race 4 and America race 1, and China race 3 and Brazil race 2 exist in similar pathopoiesia (Hu et al., 1995). Among 11 races of the fungal pathogen in China, races 1 and 7 were identified as the dominant ones, whose incidence rate were 50% and 22% respectively.
The main symptoms of FLS are small, spots like frogeye on the leaves. The infection lesions can also develop on stems, pods and seeds (Sinclair and Backman, 1989). The disease incidence favored by hot and humid environment and it brought about the reduction of the productivity derived from the reduction of the photosynthetic area, premature defoliation. Susceptible cultivars were 12%-15% in less at average case, 30% at severe case (Akem and Dashiell, 1994). Germination ratio of susceptible seeds were lower, and it’s quality fall off, 1.2% in protein, 2.9% in fat, 2g in thousand seed weight.
The utilizing natural genetic resistance and breeding resistant cultivar are the most economical and efficient means to control FLS, because the chemical control with fungicides, in addition to being expensive, hasn't been shown to be effective. In the process of the traditional resistance breeding, it was aimless to select resistant parents and segregation offspring in terms of resistance phenotype identification. Otherwise, resistance identification was expertise-required, time-consuming, laboursome and environmentally sensitive. In order to solve these problems well, DNA markers linked to resistant genes can be used. Among 11 races of C. sojina recognized in China, only the three markers Satt565, SOYGPATR, Satt396 (Zhang et al., 2004) linked with resistance gene physiological race 1 and one RAPD marker OPS036 with resistance gene against physiological race 7 (Zhou et al., 1998) were identified. These DNA markers were insufficient for assisted-select resistant breeding against FLS. The main purpose of this research was to find new molecular markers linked with resistance gene by analyzing resistance genetic and mapping the resistance locus against FLS.
1 Results
1.1 Inheritance of the resistance
After inoculation with C. sojina race 7, the FLS reaction of the two parents, ‘Gang 95144-1’ and ‘Gongjiao 9723-6’ are significantly different. The reaction of ‘Gongjiao 9723-6’ was susceptible and the reaction of ‘Gang 95144-1’ was highly resistant. Among the 184 F2 individuals inoculated, 138 individuals were resistant and 46 individuals were susceptible. A good fit to a 3:1 resistant:susceptible ratio (χ2 =1.15, P =0.868) was showed. It was indicated that the resistance in ‘Gang 95144-1’ was completely dominant and controlled by a single gene.
1.2 Identification of SSR markers
A total of 600 SSR primers were selected to survey the polymorphisms between ‘Gang 95144-1’ and ‘Gongjiao 9723-6’. 24 primer pairs couldn’t obtain amplified DNA bands, the remaining 363 primer pairs couldn’t obtain polymorphism between the two parents, and 213 primer pairs obtained polymorphic bands that ranged from 100 bp to 300 bp in size.
These polymorphic primer pairs between parents were used to detect the polymorphisms between the two DNA bulks (Resistant and susceptible). Only 4 primer pairs (Satt207, Satt411, Satt384 and Satt491) exhibited polymorphisms between the two DNA bulks. The results suggest that these markers were relevant possibly to resistant locus to Cerocospara sojina race 7 in soybean.
1.3 Linkage analysis
The polymorphism markers between the two DNA bulks were screened in the entire F2 population to analyze the resistance segregation. The χ2 test was used to analyze the genetic segregation of the 4 polymorphic primer pairs in F2 population. The χ2 value of the Satt411 and Satt384 marker were respectively 1.570 and 2.562, and the P value were respectively 0.473 and 0.317, indicating that their maternal type,heterozygous type and paternal type conformed significantly to 1:2:1 (Table 1, Figure 1 and Figure 2). The χ2 value of the Satt207 and Satt491 marker were respectively 66.687 and 334.607, and the P value were respectively 5.32×10-15 and 2.19×10-73, indicating that their maternal type, heterozygous type and paternal type did not conform to 1:2:1 (Table 1).
Figure 1 DNA amplified products for the SSR marker Satt414 Note: P1: ‘Gang95144-1’; P2: ‘Gongjiao9723-6’; R: resistant DNA bulk; S: susceptible DNA bulk; 1~30 some plants among F2 population derived from the cross of ‘Gang 95144-1’ בGongjiao 9723-6’ |
Table1 Genetic Segregation of Polymorphic SSR primer pairs between the two DNA bulk in F2 population |
Figure 2 DNA amplified products for the SSR marker Satt384 Note: P1: ‘Gang95144-1’; P2: ‘Gongjiao9723-6’; R: resistant DNA bulk; S: susceptible DNA bulk; 1~38 some plants among F2 population derived from the cross of ‘Gang 95144-1’ בGongjiao 9723-6’ |
Analyzed based on the Mapmaker3.0 software, it was found that Satt411 and Satt384 were linked with the resistant locus. The linkage order and distance was satt384 -5.7cM -satt411 -7.9cM –Hrcs7. (Figure 3) . Based on the Cregen’s genetic linkage map of soybean (Cregen, 1999), the resistant locus was located in E linkage group.
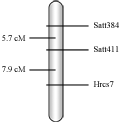 Figure 3 The position of resistant locus to soybean Cercospora leaf spot Race 7 integrated into the Cregen’s genetic linkage map of soybean |
2 Discussions
Genetics of plant-pathogen interactions specify that plants often contain single dominant resistance genes that specifically recognize pathogens that contain complementary avirulence genes (Flor, 1971). Our results showed that one major dominant gene in ‘Gang 95144-1’ confer resistance to C. sojina race 7, which was consistent with those reported on the resistance to C. sojina race 7 and race 1 in China (Yang et al.,1995; Zhou et al.,1998; Zhang et al, 2004),as well as other races in America (Athow and Probst, 1952; Philips and Boerma, 1982).
The genetic mapping result show the resistant locus was located in E linkage group and linked with satt411 and satt384, Which was different from the MLG J of American Rcs3 (Mian et al., 1999) and MLG C1 of china race 1(Zhang et al., 2004). It can be speculated that the resistant genes to the different races of C. sojina possibly distributed in different genetics linkage groups. Together with resistance controlled by one major dominant gene, we conclude molecular markers assisted-polymerizing muti races was relatively easy and was significant for the improvement of resistance to frogeye leaf spot.
MLG E was also the cluster distribution of resistant locus to soybean cyst nematode and Sclerotinia Sclerotiorum. There were respectively 3 and 2 QTL distributed in MLG E (Guo et al., 2006; Guo et al., 2008). And near the resistant locus to C. sojina race 7, there was 1 QTL of the resistance locus to soybean cyst nematode and Sclerotinia Sclerotiorum. QTL of the former were located between Satt411and Satt384 (Guo et al., 2006) and that of the later were located between Satt411 and Satt212 (Guo et al., 2008). It can be speculated that the region around Satt411 was a new cluster of resistant locus and MLG E was possiblely a main resistance linkage group to fungi.
3 Materials and Methods
3.1 Genetic materials and phenotypic assay
A mapping population of soybean was gained from a cross between the resistant ‘Gang 95144-1’ and the susceptible ‘Gongjiao 9723-6’ to C. sojina race 7. The F1 seeds were grown and selfed, and the leaf of F2 plants were used for disease evaluation and DNA extraction. The parents (‘Gang 95144-1’ and ‘Gongjiao 9723-6’) and the F2 population including 184 individuals were scored for the inoculation with race 7 of C. sojina. The inoculation of the pathogen and the evaluation of symptoms were fulfilled as described by Dong et al. (2007).
3.2 DNA bulks and PCR amplifications
Two DNA bulks (Resistant and susceptible) were produced by respectively equally pooling the DNA of 15 resistant and 15 susceptible F2 plants. The DNA extraction was carried out as described by Rogers (1998).
PCR reactions were composed of 1×PCR Buffer, 2 mmol/L MgCl2, 100 μmol/L of each dNTP, 0.4 μmol/L of each primer, 20 ng~25 ng template DNA, and 1 U of Taq DNA polymerase in a total volume of 20 μL. PCR was carried out with the conditions 94°C for 30s, 47°C for 30s, 72°C for 30s, return to step 1 35 times, 72°C for 3 min. PCR products were separated on 6% SDS-polyacrylamide gel electrophoresis and visualized by silver staining (Tiler et al., 1997)
3.3 SSR and linkage analysis
A total of 600 SSR markers were selected out of 20 genetic linkage groups (http://bldg6.-arsusda. gov/~pooley/soy/cregan/soymap.htm) every 5 cM~10 cM and tested for DNA amplification. The polymorphism markers between the ‘Gang 95144-1’ and ‘Gongjiao 9723-6 were screened against two DNA bulks (Resistant and susceptible). The polymorphism markers between the two DNA bulks were screened against the 184 F2 individuals.
The resistance trait and genetic segregation distribution of 184 F2 individuals were both detected by the Chi-square (χ2) test for goodness of fit. The data obtained from the F2 population of ‘Gang 95144-1’ × ‘Gongjiao 9723-6’ were analyzed using Mapmaker/Exp v 3.0 (Lander et al., 1987). The Kosambi map function of Mapmaker/Exp v 3.0 was used to develop a linkage map and obtain centimorgan (cM) values. The markers were assigned to linkage groups by the “group” command with a 3.0 minimum LOD and 50 cM maximum distance and then arranged by the “order” command. Basing on the published map the linkage groups were anchored to soybean chromosomes (Cregen, 1999).
Authors’ contributions
DZM and WSM analyzed the trait phenotype and the genotype data and drafted the manuscript. LJ worked on the trait phenotype identification. LZ had the experiment on identification of SSR markers. LZG performed the field experiment management. All authors read and approved the final manuscript.
Acknowledgements
We thank Dr. Liang Chen (Biotechenology Centre, Jilin Academy of Agriculture Science, Chuangchun, China) for critical reading and formatting of the manuscript, and Professor Pengyin Chen (Department of crop, soil, and enviromental sciences, University of Arkansas, Arkansas, American) for help and advice on the experiment .This study was jointly supported by Chinese National Programs for High Technology Research and Development(Grant No.2006AA100104-6 and No 2006AA10Z1F1-2).
References
Akem C.N., and Dashiell K.E., 1994, Effect of planting date on severity of frogeye leaf spot and grain yield of soybean, Crop Protect, 13: 607-610 doi:10.1016/0261-2194(94)90006-X
Athow K.L., and Probst A.H., 1952, The inheritance of resistance to frogeye leaf spot of soybean, Phytopathology, 42: 660-662
Cregen P.B., 1999, An integrated genetic linkage map of the soybean genome, Crop Science, 39: 1464-1489 doi:10.2135/cropsci1999.3951464x
Dong Z.M., Liu J., and Liu Y.Z., 2007, Resistance to race 7 of cercospora sojina hara in northern-spring soybean, Dadou Kexue (Soybean Science), 26(5): 732-735
Flor H.H., 1971, Current status of the gene-for-gene concept, Annu. Rev. Phytopathol., 9: 275-296 doi:10.1146/annurev.py.09.090171.001423
Guo B., Sleper D.A., Lu P., Shannon J.G., Nguyen H.T., and Arelli P.R., 2006, QTLs associated with resistance to soybean cyst nematode in soybean: meta-analysis of QTL locations syst nematode, Crop Science, 46(2): 595-602 doi:10.2135/cropsci2005.04-0036-2
Guo X.M., Wang D.C., and Anne E.D., 2008, Genetic mapping of QTL underlying partial resistance to sclerotinia sclerotiorum in soybean PI 391589A and PI 391589B, Crop Science, 48: 1129-1139 doi:10.2135/cropsci2007.04.0198
Hu G.H., Yu F.Y., Cheng X.W., Yong J.K., and Xin H.H., 1995, Identification of physiological race of cercospora sojina hara in soybean, Zhiwu Baohu (Plant Protection), 21(3): 26-28
Lander E.S., Green P., Abrahamson J., Barlow A., Daly M.J., Lincoln S.E., and Newburg L, 1987, MAPMAKER: an interactive computer package for constructing primary genetic maps of expermental and natural populations, Genomics, 1: 174-181
doi:10.1016/0888-7543(87)90010-3
Mian M., Wang T.Y., Phillips D.V., Alvernaz J., and Boerma H.R., 1999, Molecular Mapping of the Rcs3 gene for resistance to Frogeye Leaf Spot in Soybean, Crop Sci., 39: 1687-1691 doi:10.2135/cropsci1999.3961687x
Michelmore R.W., Paran I., and Kesseli R.V., 1991, Identification of markers linked to disease resistance genes by bulked segregate analysis: A rapid method to detect markers in specific genomic regions by using segregating populations, Proc. Natl. Acad. Sci. USA., 88: 9828-9832 doi:10.1073/pnas.88.21.9828
Philips D. V., and Boerma H.R., 1982, Two genes to resistance to race 5 of Cer cospora sojinain soybean. Phytopathology, 72: 764-766 doi:10.1094/Phyto-72-764
Rogers O.S., 1998, Extraction of DNA from plant tissues, Plant Molecular Biology, 6: 1-10
Sinclair J.B., and Backman P.A., 1989, Frogeye leaf spot, In: Sinclair JB and Shurtleff MC (eds), Compendium of soybean diseases. 3rd ed, Amer Phytopathol Soc, St. Paul, pp.19-21
Tixier M.H., Sourdille P., Roder M., Leroy P., and Bernard M., 1997, Detection of wheat microsatellites using a no radioactive silver2 nitrate staining method, Journal Genetic Breed, 51: 175- 177
Yang Q.K., Zhang X.G., and Wang J.L., 1995, Inheritance of resistance of resistance to race 7 of cersoospora soina hara, Dadou Kexue (Soybean science), 14(1): 80-82
Yorinori J.T., 1997, Potential for integrated disease management in soybean Proceedings, World Soybean Research Conference V, Chiang Mai, Kasetsart University Press, Bangkok, pp.233-238
Zhang W.H., Chen Q.S., Yang Q.K., Li W.B., Wang W.H., Liu C.Y., Chen L.J., Liu H.Y., and Shan J.X., 2004, Analysis of resistance gene against cercospora sojina hara race 1 in soybean with SSR marker. Dadou Kexue (Soybean science), 3(23): 169-173
Zhou J.J., Dong W., Yang Q.K., Cao Y.P., and Chen S.Y., 1998, Genetic analysis and RAPD marker of resistance to race 7 of cercospora sojina hara in soybean. Kexue Tongbao (Science bulletin), 21(43): 2301- 2307
. PDF(348KB)
. FPDF
. HTML
. Online fPDF
Associated material
. Readers' comments
Other articles by authors
. Zhimin Dong
. Shuming Wang
. Jia Liu
. Zhi Li
. Zhigang Yi
Related articles
. Soybean (Glycine max)
. Cercospora leaf spot Race 7
. Resistant locus
. Hrcs7
. Genetic mapping
Tools
. Email to a friend
. Post a comment


